Greetings, savvy synthesists! As of January 2026, we’ve implemented updates to our project setup coding form Wizard and streamlined the available template options to provide a faster, more flexible setup experience (see the “Setting Up a Project” section of the User Guide for more specifics). With these updates in place, we wanted to offer a refreshed overview of two key points related to utilizing the longitudinal correlational template and compatibility with other coding form structures:
- When you should choose the longitudinal correlational template (vs. another template option that allows you to code correlations as a type of effect size, but not longitudinal correlations), and
- The steps involved in setting up your project workflow when your review requires capturing both longitudinal correlations and other effect size types, such as standardized mean differences (SMDs).
You may remember our earlier blog post introducing the longitudinal correlational meta-analysis coding form template. That resource remains the best comprehensive guide for understanding the purpose of correlational meta-analyses and the specific structure of a longitudinal correlational coding form.
The purpose of the current post is to offer guidance on project setup for synthesists working with correlation coefficients, especially those focusing on syntheses of longitudinal correlations alongside other effect size types.
Longitudinal Correlational vs. Traditional Coding Forms: What’s in a (Data) Frame?
For all coding forms across various effect sizes, MetaReviewer captures and exports data using a hierarchical relational database structure, or HRD (see here in case you need a refresher!). The Effect Size page is where the magic happens: identifiers from prior pages (like sample IDs and measure IDs) “link up” to determine the unique values of each row, such that each row constitutes a unique effect size while higher-level details, such as sample and measure characteristics, are preserved. Although certain study-level characteristics that are common across effect sizes may be repeated across rows—such as the study design, study quality, and funding information—within each row, the combination of information entered on the Effect Size page is unique.
For syntheses of standardized mean differences (SMDs) within or between groups, which tend to utilize aggregate statistics such as regression coefficients, gain scores, and raw or adjusted means and standard deviations, the unique identifiers on the Effect Size page (and within each row of the export) include:
- Sample_id
- The unique ID assigned by users to define the sample being coded (e.g., IG1_female_students)
- Sample_id_2
- An exact copy of the Sample_id, which users must enter again here to facilitate linkage on the Effect Size page
- Condition_id
- The unique ID assigned by users to define the condition being coded (e.g., IG_Cond_1)
- Condition_id_2
- An exact copy of the Condition_id, which users must enter again here to facilitate linkage on the Effect Size page Measure_id
- The unique ID assigned by users to define the measure being coded (e.g., Math_Score)
- Measure_id_2
- An exact copy of the Measure_id, which users must enter again here to facilitate linkage on the Effect Size page
- EffectSizeID
- The unique ID assigned by users to define the effect size being coded (e.g., es_1, es_2). Note that the input is arbitrary but must be different across rows.
- Es_selection
- ...and individual fields reflecting the information required to compute a standardized mean difference value given the user-specified effect size type entered in “Es_selection”
For syntheses of longitudinal associations between variables, which tend to utilize aggregate statistics such as Pearson’s r and Spearman’s rho, the unique identifiers on the Effect Size page (and within each row of the export) are:
- Sample_id
- The unique ID assigned by users to define the sample being coded (e.g., IG1_female_students)
- Wave_id
- The unique ID assigned by users to define the time (or wave) at which the predictor and outcome variables represented in the correlation coefficient were collected (e.g., Wave_1_baseline, Wave_2_6mo)
- Wave_id_2
- An exact copy of the Wave_id, which users must enter again here to facilitate linkage on the Effect Size page
- Predictor_id
- The unique ID assigned by users to define the predictor measure being coded (e.g., Grade_4_Math_Score)
- Outcome_id
- The unique ID assigned by users to define the outcome measure being coded (e.g., Grade_5_Science_Score)
- Effect_size_id
- The unique ID assigned by users to define the effect size being coded (e.g., effect_1, effect_2). Note that the input is arbitrary but must be different across rows.
- Es_selection
- ...and individual fields reflecting the information required to compute a standardized correlation coefficient value given the user-specified effect size type entered in “Es_selection”
As you can see above, the SMD and correlational coding forms each capture different pieces of information and rely on distinct sets of IDs to populate the Effect Size page. For example, the longitudinal correlational coding form does require condition IDs, and the SMD coding form does not require wave IDs. As a result, MetaReviewer users cannot code SMD and longitudinal correlational effect size types within a single coding form. Even if project setup and data entry appear to proceed smoothly, the data export will be uninterpretable, with misaligned columns, transposed fields, and missing data.
Why? Because identifiers from early pages are linked on the Effect Size page and those identifiers differ between SMD and longitudinal correlational coding forms. The underlying structures of each coding form are simply not aligned. Without consistent linking IDs to connect items on the coding pages to the Effect Size page, MetaReviewer cannot merge those data structures, and fields will be vertically stacked rather than horizontally aligned upon export. The result?
- A tall, stretched, messy stack instead of a tidy, wide, analysis-ready dataset
- SMD fields appearing under correlation fields (and vice versa)
- Difficulty cleaning or reshaping the data post‑export
This misalignment isn’t a bug, it’s a feature: a direct consequence of MetaReviewer customization. Each form is encoded and linked differently, within two distinctly organized databases that cannot be combined.
What should you do if you need to code both SMDs and longitudinal correlations?
Simply create two coding forms – one for each effect size type. For example, imagine a synthesist who is conducting a meta-analytic structural equation modeling (MASEM) project. They need to code information for two sets of effect sizes: SMDs between conditions reflecting impacts of an intervention on outcomes, and longitudinal correlations reflecting a theorized mediator by which intervention impacts operate. Rather than coding everything all together, this synthesis will need to set up two coding forms: one for their SMDs, and a second for their longitudinal correlations. After coding, they will have two clean, correctly structured datasets for export, which can be merged by various combinations of identifiers for synthesis. This particular synthesist would likely merge by the unique combination of study_id, citation_id, and sample_id, which would appear in the same unique combination in both exports. This solution allows users to make the most of MetaReviewer's flexibility by combining two different hierarchical relational database structures for the same synthesis project.
- Helpful hint for MASEM projects: Be sure to rename the variables in your merged dataset to clearly distinguish between correlational and SMD measures. For example, in the example above, measure_id_1 should be called SMD_measure; measure_id_1.1 should be called corr_pred_measure; measure_id_1.2 should be called corr_outcome_measure. Your variable names may differ, so be sure to consider your particular project and datasets!
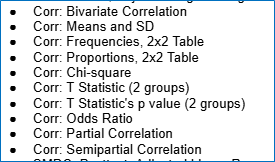
Got it: no mixing & matching! But what about the “Corr:..” Effect Size type options in the es_selection variable within SMD-focused coding forms?
These “Corr:...” effect sizes options are for cross-sectional correlations, not longitudinal correlations. They are designed to capture within- or between-group associations between variables—which can themselves be synthesized, or can be converted to a group differences metric (such as an SMD or other effect size type) for synthesis. They are not to be used for longitudinal correlations that involve multiple waves or multiple predictor/outcome pairings. Instead, they represent a single condition (and possibly a comparison) and a single measure indicator.
We hope that clears things up and helps you set up your longitudinal, correlational synthesis project for success! If things are still fuzzy, don’t hesitate to reach out to the Help Desk.



